Blossom
Link: github.com/blossom-evolution/blossom
The idea
blossom is a simple evolution software package that I began in Spring 2018. I got the idea while listening to a research talk about genetics and mutations at Harvard. At the time, I was working with astrophysics fluid dynamics simulations, so I thought: what if I wrote my own minimalistic simulation suite in which organisms could mutate?
I began reducing the idea to its absolute simplest – an organism could be represented by a list such as [dna, position, age] = ["0100", [4, 2], 3], and “mutations” could occur simply by random changing elements in the dna. Organisms would then only be able to reproduce with one another if their DNA sequences are similar enough (a very crude approximation of nature).
This idea “blossomed” into its current realization as a more detailed and careful treatment of organism evolution. Instead of simple lists, Organisms are objects that live in and interact with a World object, all of which is contained with a Universe. As I thought more and more about the possibilities of the simulation, I made sure to be very careful in establishing the framework behind the simulation package. This basically meant I made design decisions way before I coded anything up, and started with the very basics – could I successfully instantiate a universe with a population of stationary organisms with absolutely no other behaviors and read and write datasets successfully? By thinking through the basics beforehand, extending this to random movement, then to reproduction via replication, then to drinking from water located in the world, etc., was pretty straightforward!
Some of the more interesting and ambitious behaviors that I’ve played around with include movement influenced by thirst or hunger, the ability to eat other organisms, and sexual reproduction (as opposed to spontaneous replication). I made sure to allow the inclusion of external Python methods (via callbacks) so that users can write their own behaviors and take further control of each simulation.
A quest for the predator-prey model
I made it a small grail of blossom to try to replicate the prototypical predator-prey model, i.e. governed by the Lotka-Volterra equations. It felt like a good milestone because it’s simple enough to be attainable but substantial enough to be interesting.
So we need 2 species (“predator” and “prey”), custom methods that govern how the prey replicates and the predator eats, and methods that govern how each organism moves. Ultimately, it took a lot of trial and error to get something close; in the future, it’d be better to write some kind of grid search method to search for ideal simulation parameters to yield the population dynamics we’re hoping for. However, each simulation takes a good amount of time on my computer, since it iterates over every organism at every timestep. There are probably clever ways to accelerate this in the future, such as multithreading, but I want to ensure that each organism has an equal chance of acting based on its state and surroundings.
To track simulation progress in terms of species population counts, I created a local dashboard that runs in the browser using Dash and Plotly. Here’s an example of what the dashboard looks like on decent run of a predator-prey model:
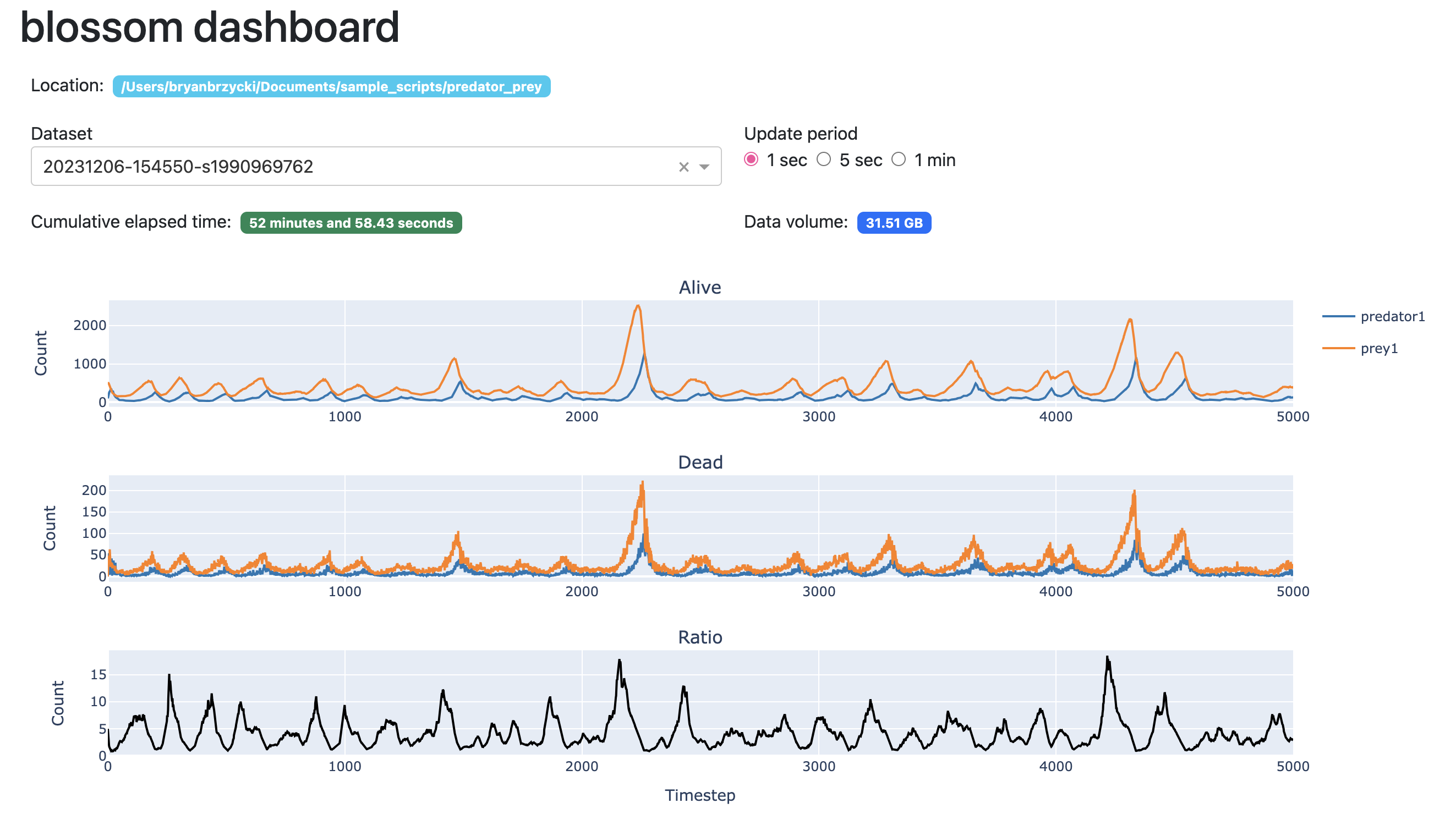
The project is located at github.com/blossom-evolution.
